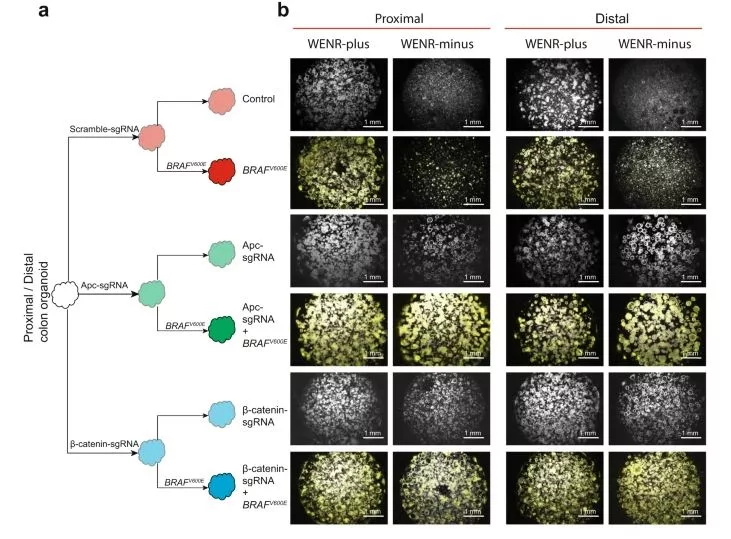
Cancers originating from the same tissue type but located in different anatomical regions often exhibit distinct molecular characteristics influencing tumorigenesis. This study focuses on proximal and distal colon cancers, highlighting the prevalence of BRAFV600E mutations in proximal colon cancers, accompanied by an increased DNA methylation phenotype. Using mouse colon organoids, the research reveals that proximal and distal colon stem cells possess unique transcriptional programs governing stemness and differentiation. The homeobox transcription factor CDX2, silenced by DNA methylation in proximal colon cancers, plays a crucial role in mediating these differential transcriptional programs. The study demonstrates that the Cdx2-mediated proximal colon-specific transcriptional program acts as a tumor suppressor. Loss of Cdx2 creates a permissive state for BRAFV600E-driven transformation. Human proximal colon cancers with downregulated CDX2 exhibit a transcriptional program similar to mouse proximal organoids with Cdx2 loss. The findings underscore the significance of developmental transcription factors, such as CDX2, in maintaining location-specific transcriptional programs that contribute to tissue-type origin-specific dependencies in tumor development.
Keywords: Organoids, oncology