RNA-Binding Proteins: Orchestrating Chromatin Architecture in Cell Differentiation
Journal: Nat Cell Biol (2025).
Author: Dehingia, B., Milewska-Puchała, M., Janowski, M. et al., Spain / USA
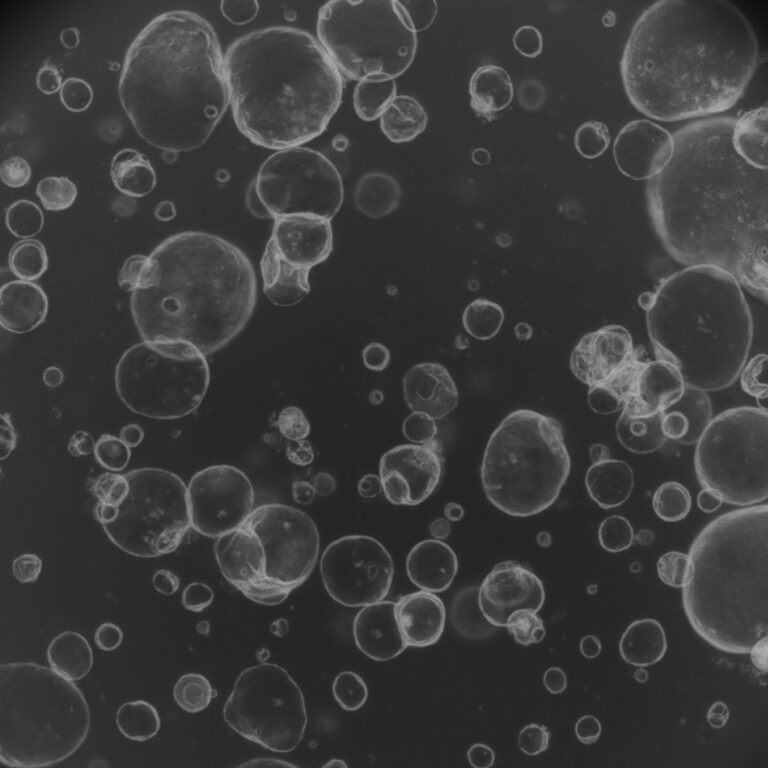
New research uncovers how RNA-binding proteins (RBPs) help shape chromatin structure during stem cell differentiation. As embryonic stem cells transition into neural stem cells, interactions between RBPs and the architectural protein CTCF increase, reinforcing chromatin loops and domain boundaries. A non-coding RNA, Pantr1, emerges as a key player, enhancing RBP – CTCF interactions and driving chromatin maturation. This structural reinforcement ensures proper gene regulation – keeping neuronal genes silenced until the right developmental stage. The study highlights a fundamental mechanism linking chromatin topology to controlled gene expression during differentiation.
Boosting CRISPR Delivery with Lipid Nanoparticle – Spherical Nucleic Acids
Journal: Proc. Natl. Acad. Sci. U.S.A. 122 (36) e2426094122, (2025)
Author: Z. Han,C. Huang,T. Luo, & C.A. Mirkin, USA
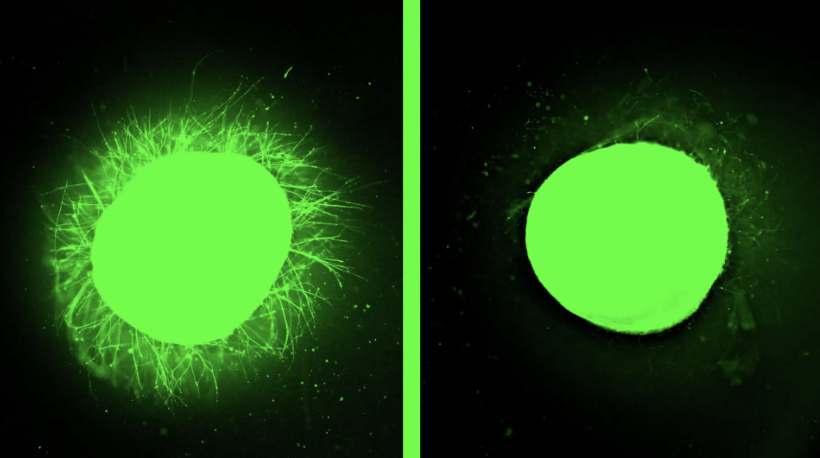
A new delivery platform combines lipid nanoparticles with spherical nucleic acids to enhance CRISPR–Cas gene editing. These CRISPR LNP–SNAs show 2–3× higher uptake, lower toxicity, and greater editing efficiency compared to standard nanoparticles. They also improve homology-directed repair, offering a safer and more versatile route for genetic therapies.
Uncovering New Vulnerabilities in Small-Cell Lung Cancer
Journal: Sig Transduct Target Ther 10, 290 (2025)
Author: Wang, GZ., Wang, Z., Bai, SH. et al., China
A large multiomics study of over 300 small-cell lung cancers (SCLC) reveals deep tumor heterogeneity and identifies frequent abnormal splicing of the focal adhesion kinase (FAK) gene. These FAK variants drive aggressive disease but also create a therapeutic vulnerability – showing strong sensitivity to FAK inhibitors in patient-derived models. The findings highlight new molecular targets for tackling this highly lethal cancer.
AI-Powered Pathology Sharpens Breast Cancer Spatial Transcriptomics
Journal: npj Precis. Onc. 9, 310 (2025)
Author: Li, T., Yang, Q., Acs, B. et al., Sweden
Researchers developed a machine learning–based computational tissue annotation (CTA) pipeline to enhance spatial transcriptomics in breast cancer. By mapping tumor, stroma, and immune compartments from pathology images, CTA improves resolution beyond current tools, offering deeper insights into tumor heterogeneity, immune interactions, and cancer subtypes. This approach could advance precision diagnostics and personalized treatment strategies.